Calculate Edu+ (ch02) cell in stack of DAPI nuclei (ch01)
Download link (.txt): wubboscript_vs12_orderC2_dapi_C1_EdU
Autogrid in ROI: measuring intensity distribution (Wubbolts 2013)
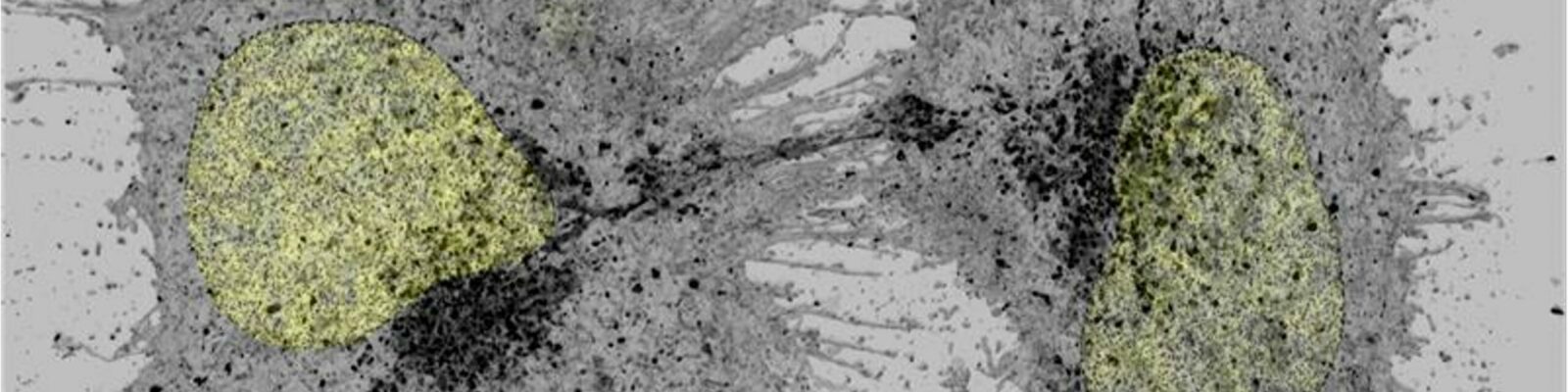
Developed to measure Edu+ cells in a DAPI stained organoids (N. Pelaez Llaneza / R.Wubbolts).
Prepare: Make stack images of organoids (ch01= dapi, ch02 =Edu), z-space 1 µm.
The .lif files should converted to –c1.tif and –c2.tif files using the excellent macro of Christoph Letterier (LIF extractor), the result will give you a list of the files within the .lif dataset split into channels (-c1.tif and –c2.tif and a rgb merge).
Does: Asks for DAPI (-C1.tif) image, opens –c2 image with title match, combines both images (EdU seems to dampen DAPI signal), then determines centroids of objects in both channels and removes those that are closer than 1 µm from the ROI manager list(dist variable) and restricts this selection to neigbouring z-slices (ds=2), number of deleted Rois are printed in the log. In addition, images in the stack are taken together (z-reduction factor) to further prevent overlapping objects. Then it measures number of objects in the total object count and Edu+ object count. Percentages are calculated and placed into a result table. A view of the stack is give and the roimanager allows toggling of the rois that were selected. This is saved in a results directory.
Options:
Vary ds and dist (Line 110)
Vary z-reduction factor (Line 20): collapses n-slices (2) to reduce oversampling of structures in the stack.
Credits: Christoph Letterier / Vytas Bindokas / Wayne Rasband / Bioformats and FIJI development teams